Writing a reproducible paper¶
This use case demonstrates how to use nested DataLad datasets to create a fully reproducible paper by linking
(different) DataLad dataset sources with
the code needed to compute results and
LaTeX files to compile the resulting paper.
The different components each exist in individual DataLad datasets and are aggregated into a single DataLad superdataset complying to the YODA principles for data analysis projects[1]. The resulting superdataset can be publicly shared, data can be obtained effortlessly on demand by anyone that has the superdataset, and results and paper can be generated and recomputed everywhere on demand.
A template to start your own reproducible paper with the same set up can be found on GitHub.
The Challenge¶
Over the past year, Steve worked on the implementation of an algorithm as a software package. For testing purposes, he used one of his own data collections, and later also included a publicly shared data collection. After completion, he continued to work on validation analyses to prove the functionality and usefulness of his software. Next to a directory in which he developed his code, and directories with data he tested his code on, he now also has other directories with different data sources used for validation analyses. “This cannot take too long!” Steve thinks optimistically when he finally sits down to write up a paper.
His scripts run his algorithm on the different data collections, create derivatives of his raw data, pretty figures, and impressive tables. Just after he hand-copies and checks the last decimal of the final result in the very last table of his manuscript, he realizes that the script specified the wrong parameter values, and all of the results need to be recomputed - and obviously updated in his manuscript. When writing the discussion, he finds a paper that reports an error in the publicly shared data collection he uses. After many more days of updating tables and fixing data columns by hand, he finally submits the paper. Trying to stand with his values of open and reproducible science, he struggles to bundle all scripts, algorithm code, and data he used in a shareable form, and frankly, with all the extra time this manuscript took him so far, he lacks motivation and time. In the end, he writes a three page long README file in his GitHub code repository, includes his email for data requests, and secretly hopes that no-one will want to recompute his results, because by now even he himself forgot which script ran on which dataset and what data was fixed in which way, or whether he was careful enough to copy all of the results correctly. In the review process, reviewer 2 demands that the figures his software produces need to get a new color scheme, which requires updates in his software package, and more recomputations.
The DataLad Approach¶
Steve sets up a DataLad dataset and calls it algorithm-paper. In this
dataset, he creates several subdirectories to collate everything that is relevant for
the manuscript: Data, code, a manuscript backbone without results.
code/ contains a Python script that he uses for validation analyses, and
prior to computing results, the script
attempts to download the data should the files need to be obtained using DataLad’s Python API.
data/ contains a separate DataLad subdataset for every dataset he uses. An
algorithm/ directory is a DataLad dataset containing a clone of his software repository,
and within it, in the directory test/data/, are additional DataLad subdatasets that
contain the data he used for testing.
Lastly, the DataLad superdataset contains a LaTeX .tex file with the text of the manuscript.
When everything is set up, a single command line call triggers (optional) data retrieval
from GitHub repositories of the datasets, computation of
results and figures, automatic embedding of results and figures into his manuscript
upon computation, and PDF compiling.
When he notices the error in his script, his manuscript is recompiled and updated
with a single command line call, and when he learns about the data error,
he updates the respective DataLad dataset
to the fixed state while preserving the history of the data repository.
He makes his superdataset a public repository on GitHub, and anyone who clones it can obtain the
data automatically and recompute and recompile the full manuscript with all results.
Steve never had more confidence in his research results and proudly submits his manuscript.
During review, the color scheme update in his algorithm sourcecode is integrated with a simple
update of the algorithm/ subdataset, and upon command-line invocation his manuscript updates
itself with the new figures.
Take a look at the real manuscript dataset
The actual manuscript this use case is based on can be found
here:
https://github.com/psychoinformatics-de/paper-remodnav. datalad clone (manual)
the repository and follow the few instructions in the README to experience the
DataLad approach described above.
There is also a slimmed down template that uses the analysis demonstrated in YODA-compliant data analysis projects and packages it up into a reproducible paper using the same tools: github.com/datalad-handbook/repro-paper-sketch.
Step-by-Step¶
datalad create (manual) a DataLad dataset. In this example, it is named “algorithm-paper”,
and datalad create uses the yoda procedure[1] to apply useful configurations
for a data analysis project:
$ datalad create -c yoda algorithm-paper
[INFO ] Creating a new annex repo at /home/adina/repos/testing/algorithm-paper
create(ok): /home/adina/repos/testing/algorithm-paper (dataset)
This newly created directory already has a code/ directory that will be tracked with Git
and some README.md and CHANGELOG.md files
thanks to the yoda procedure applied above. Additionally, create a subdirectory data/ within
the dataset. This project thus already has a comprehensible structure:
$ cd algorithm-paper
$ mkdir data
# You can checkout the directory structure with the tree command
$ tree
algorithm-paper
├── CHANGELOG.md
├── code
│ └── README.md
├── data
└── README.md
All of your analyses scripts should live in the code/ directory, and all input data should
live in the data/ directory.
To populate the DataLad dataset, add all the
data collections you want to perform analyses on as individual DataLad subdatasets within
data/.
In this example, all data collections are already DataLad datasets or git repositories and hosted on GitHub.
datalad clone therefore installs them as subdatasets, with -d ../
registering them as subdatasets to the superdataset[2].
$ cd data
# clone existing git repositories with data (-s specifies the source, in this case, GitHub repositories)
# -d points to the root of the superdataset
datalad clone -d ../ https://github.com/psychoinformatics-de/studyforrest-data-phase2.git
[INFO ] Cloning https://github.com/psychoinformatics-de/studyforrest-data-phase2.git [1 other candidates] into '/home/adina/repos/testing/algorithm-paper/data/raw_eyegaze'
install(ok): /home/adina/repos/testing/algorithm-paper/data/raw_eyegaze (dataset)
$ datalad clone -d ../ git@github.com:psychoinformatics-de/studyforrest-data-eyemovementlabels.git
[INFO ] Cloning git@github.com:psychoinformatics-de/studyforrest-data-eyemovementlabels.git into '/home/adina/repos/testing/algorithm-paper/data/studyforrest-data-eyemovementlabels'
Cloning (compressing objects): 45% 1.80k/4.00k [00:01<00:01, 1.29k objects/s
[...]
Any script we need for the analysis should live inside code/. During script writing, save any changes
to you want to record in your history with datalad save (manual).
The eventual outcome of this work is a GitHub repository that anyone can use to get the data and recompute all results when running the script after cloning and setting up the necessary software. This requires minor preparation:
The final analysis should be able to run on anyone’s file system. It is therefore important to reference datafiles with the scripts in
code/as relative paths instead of hard-coding absolute paths.After cloning the
algorithm-paperrepository, data files are not yet present locally. To spare users the work of a manualdatalad get(manual), you can have your script take care of data retrieval via DataLad’s Python API.
These two preparations can be seen in this excerpt from the Python script:
# import DataLad's API
from datalad.api import get
# note that the datapath is relative
datapath = op.join('data',
'studyforrest-data-eyemovementlabels',
'sub*',
'*run-2*.tsv')
data = sorted(glob(datapath))
# this will get the data if it is not yet retrieved
get(dataset='.', path=data)
Lastly, datalad clone the software repository as a subdataset in the
root of the superdataset[3].
# in the root of ``algorithm-paper`` run
$ datalad clone -d . git@github.com:psychoinformatics-de/remodnav.git
This repository has also subdatasets in which the datasets used for testing live (tests/data/):
$ tree
[...]
| ├── remodnav
│ ├── clf.py
│ ├── __init__.py
│ ├── __main__.py
│ └── tests
│ ├── data
│ │ ├── anderson_etal
│ │ └── studyforrest
At this stage, a public algorithm-paper repository shares code and data, and changes to any
dataset can easily be handled by updating the respective subdataset.
This already is a big leap towards open and reproducible science. Thanks to DataLad, code,
data, and the history of all code and data are easily shared - with exact versions of all
components and bound together in a single, fully tracked research object.
By making use of the Python API of DataLad and relative paths in scripts,
data retrieval is automated, and scripts can run on any other computer.
Automation with existing tools¶
To go beyond that and include freshly computed results in a manuscript on the fly does not
require DataLad anymore, only some understanding of Python, LaTeX, and Makefiles. As with most things,
its a surprisingly simple challenge if one has just seen how to do it once.
This last section will therefore outline how to compile the results into a PDF manuscript and
automate this process.
In principle, the challenge boils down to:
have the script output results (only requires
print()statements)capture these results automatically (done with a single line of Unix commands)
embed the captured results in the PDF (done with one line in the
.texfile and some clever referencing)automate as much as possible to keep it as simple as possible (done with a Makefile)
That does not sound too bad, does it?
Let’s start by revealing how this magic trick works. Everything relies on printing
the results in the form of user-defined LaTeX definitions (using the \newcommand
command), referencing those definitions in your manuscript where the
results should end up, and bind the \newcommands as \input{} to your .tex
file. But lets get there in small steps.
First, if you want to read up on the \newcommand, please see
its documentation.
The command syntax looks like this:
\newcommand{\name}[num]{definition}
What we want to do, expressed in the most human-readable form, is this:
\newcommand{\Table1Cell1Row1}{0.67}
where 0.67 would be a single result computed by your script.
This requires print() statements that look like this in the most simple
form (excerpt from script):
print('\\newcommand{\\maxmclf}{{%.2f}}' % max_mclf)
where max_mclf is a variable that stores the value of one computation.
Tables and references to results within the .tex files then do not contain the
specific value 0.67 (this value would change if the data changes, or other parameters),
but \maxmclf (and similar, unique names for other results).
For full tables, one can come up with naming schemes that make it easy
to fill tables with unique names with minimal work, for example like this (excerpt):
\begin{table}[tbp]
\caption{Cohen's Kappa reliability between human coders (MN, RA),
and \remodnav\ (AL) with each of the human coders.
}
\label{tab:kappa}
\begin{tabular*}{0.5\textwidth}{c @{\extracolsep{\fill}}llll}
\textbf {Fixations} & & \\
\hline\noalign{\smallskip}
Comparison & Images & Dots \\
\noalign{\smallskip}\hline\noalign{\smallskip}
MN versus RA & \kappaRAMNimgFix & \kappaRAMNdotsFix \\
AL versus RA & \kappaALRAimgFix & \kappaALRAdotsFix \\
AL versus MN & \kappaALMNimgFix & \kappaALMNdotsFix \\
\noalign{\smallskip}
\textbf{Saccades} & & \\
\hline\noalign{\smallskip}
Comparison & Images & Dots \\
\noalign{\smallskip}\hline\noalign{\smallskip}
MN versus RA & \kappaRAMNimgSac & \kappaRAMNdotsSac \\
AL versus RA & \kappaALRAimgSac & \kappaALRAdotsSac \\
AL versus MN & \kappaALMNimgSac & \kappaALMNdotsSac \\
\noalign{\smallskip}
% [..] more content omitted
\end{tabular*}
\end{table}
Without diving into the context of the paper, this table contains results for three
three comparisons (“MN versus RA”, “AL versus RA”, “AL versus MN”), for three
event types (Fixations, Saccades, and post-saccadic oscillations (PSO)), and three different
stimulus types (Images, Dots, and Videos). The latter event and stimulus are omitted for
better readability of the .tex excerpt. Here is how this table looks like in the manuscript
(cropped to match the .tex snippet):
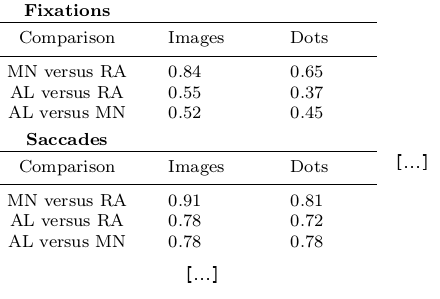
It might appear tedious to write scripts that output results for such tables with individual names.
However, print() statements to fill those tables can utilize Pythons string concatenation methods
and loops to keep the code within a few lines for a full table, such as
# iterate over stimulus categories
for stim in ['img', 'dots', 'video']:
# iterate over event categories
for ev in ['Fix', 'Sac', 'PSO']:
[...]
# create the combinations
for rating, comb in [('RAMN', [RA_res_flat, MN_res_flat]),
('ALRA', [RA_res_flat, AL_res_flat]),
('ALMN', [MN_res_flat, AL_res_flat])]:
kappa = cohen_kappa_score(comb[0], comb[1])
label = 'kappa{}{}{}'.format(rating, stim, ev)
# print the result
print('\\newcommand{\\%s}{%s}' % (label, '%.2f' % kappa))
Running the python script will hence print plenty of LaTeX commands to your screen (try it out in the actual manuscript, if you want!). This was step number 1 of 4.
How about figures?
To include figures, the figures just need to be saved into a dedicated location (for example,
a directory img/) and included into the .tex file with standard LaTeX syntax.
Larger figures with subfigures can be created by combining several figures:
\begin{figure*}[tbp]
\includegraphics[trim=0 8mm 3mm 0,clip,width=.5\textwidth]{img/mainseq_lab}
\includegraphics[trim=8mm 8mm 0 0,clip,width=.5\textwidth-3.3mm]{img/mainseq_sub_lab} \\
\includegraphics[trim=0 0 3mm 0,clip,width=.5\textwidth]{img/mainseq_mri}
\includegraphics[trim=8mm 0 0 0,clip,width=.5\textwidth-3.3mm]{img/mainseq_sub_mri}
\caption{Main sequence of eye movement events during one 15 minute sequence of
the movie (segment 2) for lab (top), and MRI participants (bottom). Data
across all participants per dataset is shown on the left, and data for a single
exemplary participant on the right.}
\label{fig:overallComp}
\end{figure*}
This figure looks like this in the manuscript:
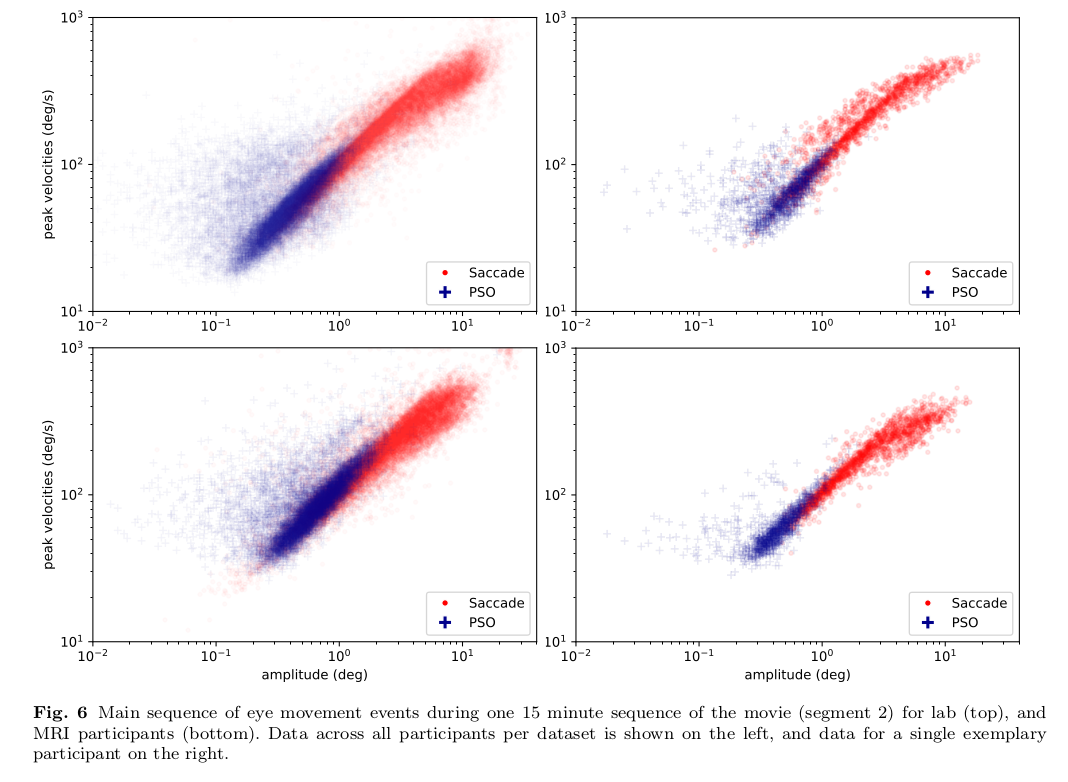
For step 2 and 3, the print statements need to be captured and bound to the .tex file.
The tee command can write all of the output to
a file (called results_def.tex):
code/mk_figuresnstats.py -s | tee results_def.tex
This will redirect every print statement the script wrote to the terminal into a file called
results_def.tex. This file will hence be full of \newcommand definitions that contain
the results of the computations.
For step 3, one can include this file as an input source into the .tex file with
\begin{document}
\input{results_def.tex}
Upon compilation of the .tex file into a PDF, the results of the
computations captured with \newcommand definitions are inserted into the respective part
of the manuscript.
The last step is to automate this procedure. So far, the script would need to be executed
with a command line call, and the PDF compilation would require another commandline call.
One way to automate this process are Makefiles.
make is a decades-old tool known to many and bears the important advantage that is will
deliver results regardless of what actually needs to be done with a single make call –
whether it is executing a Python script, running bash commands, or rendering figures, or all of this.
Here is the one used for the manuscript:
1all: main.pdf
2
3main.pdf: main.tex tools.bib EyeGaze.bib results_def.tex figures
4 latexmk -pdf -g $<
5
6results_def.tex: code/mk_figuresnstats.py
7 bash -c 'set -o pipefail; code/mk_figuresnstats.py -s | tee results_def.tex'
8
9figures: figures-stamp
10
11figures-stamp: code/mk_figuresnstats.py
12 code/mk_figuresnstats.py -f -r -m
13 $(MAKE) -C img
14 touch $@
15
16clean:
17 rm -f main.bbl main.aux main.blg main.log main.out main.pdf main.tdo main.fls main.fdb_latexmk example.eps img/*eps-converted-to.pdf texput.log results_def.tex figures-stamp
18 $(MAKE) -C img clean
One can read a Makefile as a recipe:
Line 1: “The overall target should be
main.pdf(the final PDF of the manuscript).”Line 2-3: “To make the target
main.pdf, the following files are required:main.tex(the manuscript’s.texfile),tools.bib&EyeGaze.bib(bibliography files),results_def.tex(the results definitions), and figures (a section not covered here, about rendering figures with inkscape prior to including them in the manuscript). If all of these files are present, the targetmain.pdfcan be made by running the commandlatexmk -pdf -g”Line 5-6: “To make the target
results_def.tex, the scriptcode/mk_figuresnstats.pyis required. If the file is present, the targetresults_def.texcan be made by running the commandbash -c 'set -o pipefail; code/mk_figuresnstats.py -s | tee results_def.tex'”
This triggers the execution of the script, collection of results in results_def.tex, and PDF
compilation upon typing make.
The last three lines define that a make clean removes all computed files, and also all
images.
Finally, by wrapping make in a datalad run (manual) command, the computation of results
and compiling of the manuscript with all generated output can be written to the history of
the superdataset. datalad run make will thus capture all provenance for the results
and the final PDF.
Thus, by using DataLad and its Python API, a few clever Unix and LaTeX tricks,
and Makefiles, anyone can create a reproducible paper. This saves time, increases your own
trust in the results, and helps to make a more convincing case with your research.
If you have not yet, but are curious, checkout the
manuscript this use case is based on.
Any questions can be asked by opening an issue.
Footnotes